Previous page Next page
2.3 Properties of human IgG subclasses
In the 1960's extensive studies, performed with specific polyclonal rabbit antisera against homogeneous human IgG myeloma proteins revealed the existence of four distinct subgroups of human IgG, which were designated IgG1, IgG2, IgG3 and IgG4, respectively (11,12,13). Since early studies were performed with polyclonal antisera, rendered specific by absorption, the amounts of specific reagents were relatively limited (14). In the 1980's, monoclonal antibodies against human IgG and its subclasses became available, which permitted more reproducible measurements and provided an improvement to studies dealing with IgG subclass levels in a variety of conditions (15,16,17).
Quantitatively, the relative serum concentrations of the human IgG subclasses are as follows (18,19): IgG1 > IgG2 > IgG3 = IgG4. The four subclasses show more than 95% homology in the amino acid sequences of the constant domains of the y-heavy chains. The four IgG subclasses show their most conspicuous differences in the amino acid composition and structure of the 'hinge region', which is the part of the molecule containing disulfide bonds between the y-heavy chains (figure 2). This region, between the Fab arms (Fragment antigen binding) and the two carboxy-terminal domains CH2 and CH3) of both heavy chains, determines the flexibility of the molecule (20,21). In the schematic structure of IgG, shown in figure 4, it is seen that the molecule contains domain-like structures, in which the two identical antigen-binding Fab fragments and the single Fc fragment (Fragment crystallisable) are quite mobile.
The upper hinge (towards the amino-terminal) segment allows variability of the angle between the Fab arms (Fab-Fab flexibility) as well as rotational flexibility of each individual Fab. The flexibility of the lower hinge region (towards the carboxy-terminal) directly determines the position of the Fab-arms relative to the Fc region (Fab-Fc flexibility). Hinge-dependent Fab-Fab and Fab-Fc flexibility may be important in triggering further effector functions such as complement activation and Fc receptor binding.
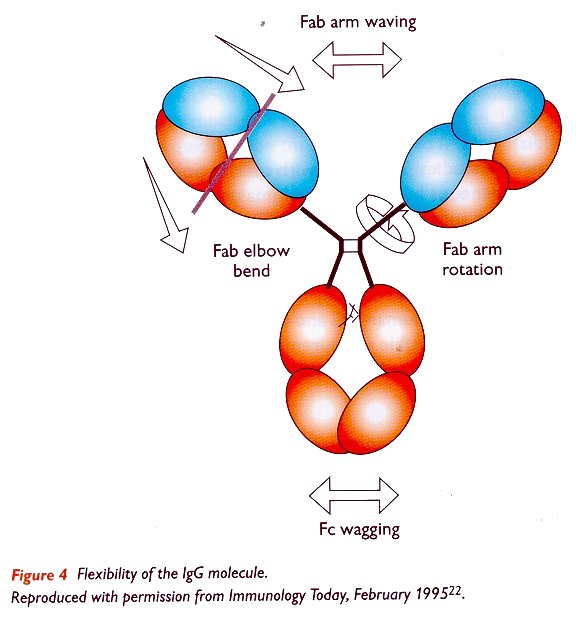
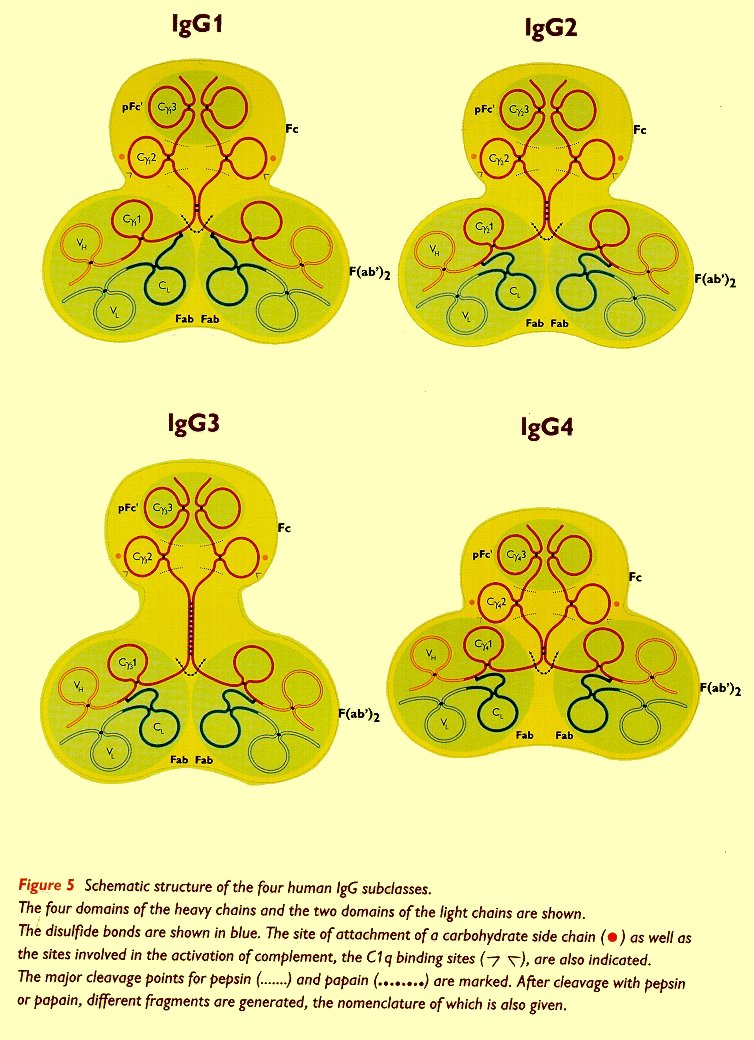
The length and flexibility of the hinge region varies among the IgG subclasses. The hinge region of IgG1 encompasses amino acids 216-231 and since it is freely flexible, the Fab fragments can rotate about their axes of symmetry and move within a sphere centered at the first of two inter-heavy chain disulfide bridges (23). IgG2 has a shorter hinge than IgG1, with 12 amino acid residues and four disulfide bridges. The hinge region of IgG2 lacks a glycine residue, it is relatively short and contains a rigid poly-proline double helix, stabilised by extra inter-heavy chain disulfide bridges. These properties restrict the flexibility of the IgG2 molecule (24). IgG3 differs from the other subclasses by its unique extended hinge region (about four times as long as the IgG1 hinge), containing 62 amino acids (including 21 prolines and 11 cysteines), forming an inflexible poly-proline double helix (25,26). In IgG3 the Fab fragments are relatively far away from the Fc fragment, giving the molecule a greater flexibility. The elongated hinge in IgG3 is also responsible for its higher molecular weight compared to the other subclasses. The hinge region of IgG4 is shorter than that of IgG1 and its flexibility is intermediate between that of IgG1 and IgG2. The schematic structure of the four IgG subclasses is shown in figure 5 (bottom of page).
As indicated in table 1, the four IgG subclasses differ with respect to the number of inter-heavy chain disulfide bonds in the hinge region (26). The structural differences between the IgG subclasses are also reflected in their susceptibility to proteolytic enzymes, such as papain (27), plasmin (28), trypsin (29) and pepsin (30).
IgG3 is very susceptible to cleavage by these enzymes, whereas IgG2 is relatively resistant. IgG1 and IgG4 exhibit an intermediary sensitivity, depending upon the enzyme used. Since these proteolytic enzymes all cleave IgG molecules near or within the hinge region, it is likely that the high sensitivity of IgG3 to enzyme digestion is related to its accessible hinge.
Another structural difference between the human IgG subclasses is the linkage of the heavy and light chain by a disulfide bond. This bond links the carboxy-terminal of the light chain with the cysteine residue at position 220 (in IgG) or at position 131 (in IgG2, IgG3 and IgG4) of the CH1 sequence of the heavy chain. Because in the folded structure these positions are close in space, they conserve the essential structure of the molecule.
In addition to differences among genes encoding the IgG subclass proteins, each with different amino acid composition and derived properties, mutations within these genes have led to variations of the composition of IgG subclass proteins within the population. The latter mutations provide the basis for genetic markers (called Gm allotypes) and correspond with minor differences in primary amino acid sequence between molecules of one IgG subclass that occur throughout a species. These allotypic determinants are polymorphic epitopes, which are inherited in a Mendelian pattern. Among individuals, different allelic forms are expressed. At present, immunoglobulin G can be typed for 18 different allotypes, located on the heavy chain. Subclass IgG3 is the most polymorphic, with thirteen Gm3 allotypes (31). There are four IgG1 allotypes and one IgG2 allotype, whereas no allotypes have been detected on the heavy chains of IgG4.Allotyping of immunoglobulins can be of diagnostic use in family and parenthood investigations (32,33).
As a consequence of the structural differences, the four IgG subclasses show differences in some of their physicochemical characteristics (table) and biological properties (table II).
TABLE I: Physiochemical properties of human IgG subclasses
|
|
IgG1 |
IgG2 |
IgG3 |
IgG4 |
|
Heavy chain type |
gamma 1 |
gamma 2 |
gamma 3 |
gamma 4 |
|
Molecular mass (kD) |
146 |
146 |
170 |
146 |
|
Amino acids in hinge region |
15 |
12 |
62 |
12 |
|
Inter-heavy chain disulfide bonds (in hinge region) |
2 |
4 |
11 |
2 |
|
Susceptibility to proteolytic enzymes |
++ |
+/- |
+++ |
+ |
|
Number of allotypes |
4 |
1 |
13 |
0 |
TABLE II: Biological properties of human IgG subclasses
|
|
IgG1 |
IgG2 |
IgG3 |
IgG4 |
|
Adult serum level range (g/l)
(mean, g/l) |
4.9-11.4
(6.98) |
1.5-6.4
(3.8) |
0.20-1.10
(0.51) |
0.08-1.40
(0.56) |
|
Proportion of total IgG (%) |
43-75 |
16-48 |
1.7-7.5 |
0.8-11.7 |
|
Half-life (days) |
21 |
21 |
7 |
21 |
|
Placental transfer |
+ |
+ |
+ |
+ |
|
Antibody response to: |
|
|
|
|
|
proteins |
++ |
+/- |
++ |
+/- |
|
polysaccharides |
+ |
++ |
(-) |
(-) |
|
allergens |
+ |
(-) |
(-) |
++ |
|
Complement activation |
|
|
|
|
|
C1q binding |
++ |
+ |
+++ |
- |
|
C1q binding, high epitope density enhancement alternative pathway |
- |
+ |
- |
+/- |
|
Binding to Fcg receptors: |
|
|
|
|
|
Fcg RI (CD64:monocytes, macrophages, neutrophils, dendritic cells) |
++ |
- |
+++ |
+ |
|
Fcg RII (CD32):monocytes, macrophages, neutrophils, eosinophils, platelets, B cells, dendritic cells, endothelial cells) |
++ |
(a) |
+++ |
- |
|
Fcg RIIa-H131 |
++ |
+++ |
+++ |
- |
|
Fcg RIIa-R131 |
++ |
- |
++ |
- |
|
Fcg RIII (CD16:neutrophils, eosinophils, macrophages, NK cells, subsets of T cells) |
++ |
- |
++ |
- |
|
Fcg RIIIb-NA1 |
+++ |
- |
+++ |
- |
|
Fcg RIIIb-NA2 |
++ |
- |
++ |
- |
|
Binding to Staphylococcal protein A |
++ |
++ |
(b) |
+ |
|
Binding to Streptococcal protein G |
++ |
++ |
++ |
++ |
|
|
|
|
|
|
|
(a):Fcg RII allotype dependent ; (b): IgG3 allotype dependent |
|
|
|
|
Previous page Next page

